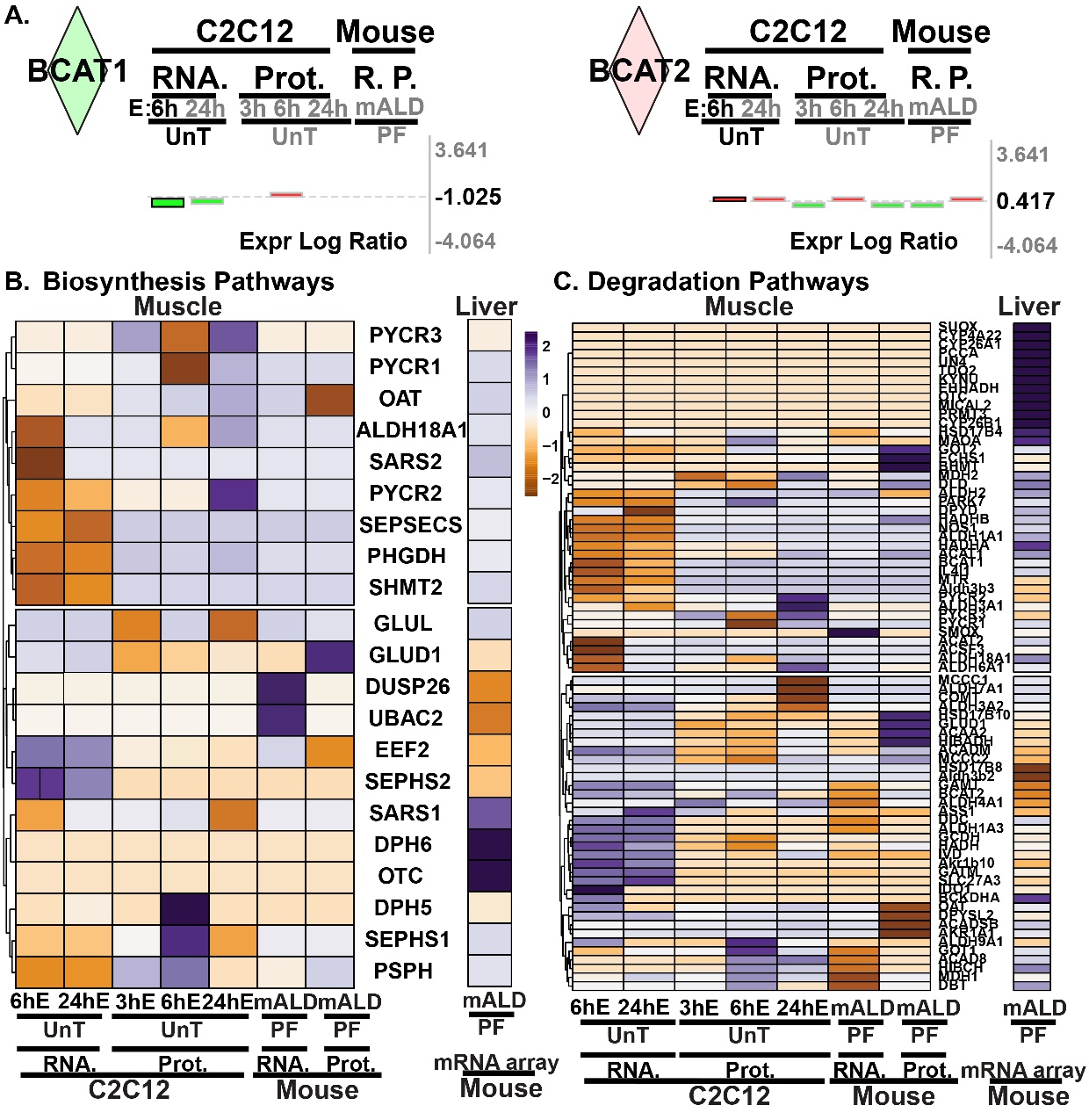
Fig. 4. Ethanol induced alterations in amino acid metabolism enzyme expression. (A). Log fold change expression levels of BCAT1 and BCAT2 in the transcriptome and proteome in C2C12 myotubes and mALD versus untreated controls and PF mice. (B). Heatmap of differentially expressed genes regulating non-essential amino acid biosynthesis pathways. (C). Heatmap of differentially expressed genes regulating proteinogenic (essential and non-essential) amino acid degradation pathways. EtOH/E: ethanol; HMB: β-hydroxy-β-methyl butyrate; UnT: untreated controls; PF: pair-fed mice; mALD: mouse model of ALD; Prot, P.: proteomics; RNA, R.: RNAseq. N = 3 biological replicates for untreated and ethanol-treated myotubes; N = 4 mice in mALD and PF groups. Significance for proteomics and transcriptomics expression levels for cells and tissue (muscle and liver) was limited to p<0.05. Significance for pathway enrichment analysis was limited to -log(p-value)= > 1.3.